Zenome Network: Distributed Genomic Internet


Abstract
Genomics is a technology spreading to many areas of human activity. Today, it is widely used in medicine, biotechnology, agriculture, and the food industry. Genetic data grows exponentially, which can make genomics the primary generator of data on the Internet (more than Twitter and YouTube). The rapid development of genomics creates several problems that need to be solved: data storage, privacy protection, the possibility of conducting large-scale scientific research, and data sharing between different organizations and people.
We believe that the development of a distributed IT infrastructure based on a peer-to-peer network and distributed ledger technology (DLT) will solve the problems mentioned above.
In this article, we present a short description for a distributed Zenome Network that is based on a new stack of protocols needed to implement a genomic IT infrastructure. Zenome Network can potentially become the foundation for the development of a new genomic Internet.
At present, there are many companies developing services using DLT in genomics: Nebula Genomics, Luna DNA, and others. Zenome Network will become a platform (new Internet) for such companies on a fundamental level of network interactions. In addition, this network will be useful for conducting cooperative genomic associative research (GWAS) by various scientific organizations without data copying.
Introduction
Many companies assert that they create solutions in the field of decentralized or new Internet [1]. In the above list there are solutions in various areas: file storage, distributed computing, messengers, social networks, and other fields [1]. It should be noted the vast majority of these projects do not have a working product or do not exist currently. Thus, despite the apparent diversity of distributed networks, the development of a new protocols stack and their implementation into a real product is still a serious and challenging task.
Our project works with genomic data and the corresponding meta-information, which imposes a number of requirements for infrastructure: (a) support for distributed data storage; (b) encryption of data when performing statistical operations (since genomic data is sensitive, they cannot be used for calculations in the natural form); (c) the ability to send messages and requests between nodes; and (d) the creation of a system of domain names. In short, we needed to reinvent the Internet in such a way that our genomic ecosystem could exist in it and users could operate with large amounts of personal data with complete security.
Since we published our Zenome whitepaper a year ago, several companies have appeared, claiming that they want to create a “blockchain-based solution” for genomics. However, most of these companies either do not have real technical documentation (Luna DNA [2], Encrypgen [3]), or the documentation shows a lack of understanding of how the claimed theses will be realized (Shivom [4], Genomes.io [5]), or companies will build a centralized solution using existing infrastructure and protocols that are not applicable to genomics (Nebula Genomics) without explaining how this will be implemented on the fundamental network level [6].
Since we first described the concept of Genomics 4.0 (similar to Industry 4.0), the basic principles of which are: Privacy, Data Sharing, Public Data Access, Right to Own Genomic Data, and a Distributed and Decentralized Environment [7], we are fully confident that the realization of our concept is possible only if there is a new Distributed Genome Internet, which will become the infrastructure for the creation of a new market for personal genomics.
In this article, we describe the main characteristics of Zenome Network, thus responding to the question of how we differ from other projects in this area — we are creating a fundamental platform for any application, enabling them to be technically implemented in a fully decentralized environment.
The article will be published shortly before the release of the beta-version of our product, so we discuss some key points of the new Zenome Network protocol stack, namely: (a) protocol development goals; (b) the general scheme and terminology of the network; ( c) describing the main network interactions of peers; (d) explaining in detail the software the architecture of the node, as the main distinguishing feature of our protocols stack, allowing the creation of a new genomic Internet, and (e) discussing how the network consensus is achieved and how we solve the problem of network scaling.
The goals of developing Zenome network protocol stack
Our goals for Zenome Network specification and its implementations are:
● Enable the use of various protocols:
○ Transports: Transmission Control Protocol (TCP), User Datagram Protocol (UDP), Datagram Congestion Control Protocol (DCCP), Stream Control Transmission Protocol (SCTP), etc.
○ Authenticated transports: Transport Layer Security (TLS), Secure Shell (SSH) Datagram Transport Layer Security (DTLS)
● Effective usage of sockets (reuse of connections)
● Enable communications between peers to be multiplexed over one socket (avoiding handshake overhead)
● Enable multi-protocols and respective versions to be used between peers, by using a negotiation process
● Backward/Forward compatibility
● Use the full capabilities of current network technologies
● Have NAT traversal
● Relayed connection possibilities
● Enable encrypted channels
● Effective usage of underlying transports
● The possibility of storage, sharing, and statistical operations on genomic data
● Implementation of distributed ledger technologies and capabilities of financial transactions
● High network stability and scalability
The general scheme of the network
To better understand the essence of network protocols, let’s look at an abstract example about Alice and Bob. Alice and Bob live in the early 20th century and communicate with each other by letters. This is during the First World War; Alice lives in Germany, and Bob lives in England; the two countries are at war with each other.
Christmas comes, and Bob wants to send Alice a Christmas card. What does Bob need for this?
Bob needs paper and a pen — that is, an interface allowing him to write the text for the letter. Next, Bob needs to write down the exact address and Alice’s full name to identify her, so that the letter will be delivered to the correct Alice. Also, there is a war, and Bob does not want intelligence to open and read his message or change the information inside the letter. Bob and Alice agree to use an Elven language unknown to the intelligence of both countries and agree to sign a note in the upper left corner. The absence of this signature is a signal that mail or the intelligence officers replaced the note.
Bob signs an envelope, pastes a postage stamp on it and sends a letter. The letter goes to the post office for sorting and is sent to different endpoints. Fortunately, the letter reaches Germany in the city where Alice lives and is again sorted into the local post office falling into the postman’s bag, which brings a letter to Alice’s house. But the trouble is, Alice’s father is a military man and was sent on a military mission to another city, and Alice no longer lives at the specified address. However, Alice’s neighbor from next door tells the postman that she knows the new address of Alice’s family and promises that she will forward the letter. And again, the letter from the neighbor goes on a journey through several sorting points and finally falls into the hands of Alice. First, she opens the letter and checks if there is a signature on the card. There is a signature and Alice is glad that she received the unaltered letter from Bob. Now Alice writes a letter to Bob and sends it, adhering to their rules. From the moment Bob receives a new address for Alice, they established regular mail communication even though their countries are at war.
So, let’s summarize what was needed to send one letter:
- Address of the recipient
- The note
- Envelope with a postage stamp
- Signature on the note
- The presence of a working postal service
- Sorting centers that distribute correspondence
- The Postman, who delivers letters
- A good neighbor who knows Alice’s new address
- Pen and paper to write a letter
But what if Bob and Alice live in 2018 and conducted joint genomic research and would like to share genetic data with privacy protection and do not want to disclose private information to a third party and use centralized servers of such companies like Google? How can they create communication and exchange data in this way?
In 2018, they can simply register on the Zenome Network and begin interaction and data exchange through the web client of the decentralized network.
In network terms, they would be peers in a distributed network, and they would need to solve the following tasks (develop the following protocols):
- Peer Identification
- Peer Discovering and Handshaking
- Peer-to-Peer Communication
- Messaging
- Routing and Switching
- Data Transfer Streams
- User Interface
Below are the main terms to give a better understanding of the operation of the distributed network.
Zenome Network Vocabulary
- Node — — a running piece of software that enables users or other programs to access network.
- Peer — — basically, a node. Two nodes of the same network are called peers.
- Module — — an interchangeable component of node, that follows the corresponding API.
- Message — — a piece of data, that goes through the network from one point to another.
- Event — — a piece of data, that’s delivered to many end points.
- Pipeline — — a number of transformers or handlers that got sequentially executed upon pieces of data flowing through pipeline.
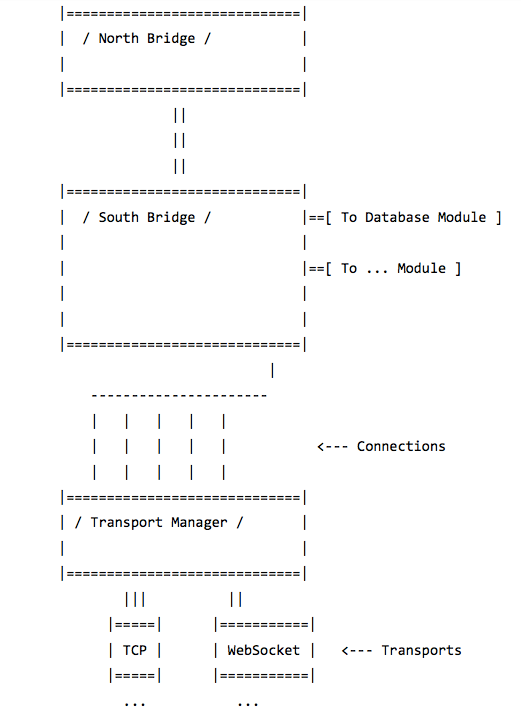
- South/North Bridge — — two distinct parts of a node (named after the corresponding parts of PC’s motherboard). South Bridge is mainly concerned with input/output operations, memory management and the network connections, while North Bridge provides interfaces and handles interactions with user tasks and services running on a node
Zenome Network Description
At Zenome project, we aim to create an extensible network suitable for storing and processing genetic information in a distributed computational process involving many nodes at the same time. In addition, there will be many services that should be running on the network, making it a market of genetic information with its own ecosystem and rich token economics (tokenomics).
Designing a distributed system is not easy and requires a lot of research and development. Some issues are so complex that they would require a book to elaborate on them. In this article, we will go another way. We will explain our current vision on the architecture of the system as simple as possible, leaving many important questions out of the scope.
First, a few words about programming languages. The initial implementation that we are working on now runs on NodeJS, and we use TypeScript to avoid the shortcomings of plain JavaScript. The same code will run everywhere, including in a browser, with some limitations.
I. The Node: Modular Configuration
The node software for our network is highly configurable. Users may, based on their needs, exclude modules that won’t be used or include experimental modules for beta testing.
For example, the build for a storage node should include an adaptor for a database and may lack computational modules or end-user graphic interfaces (top of Figure 1).
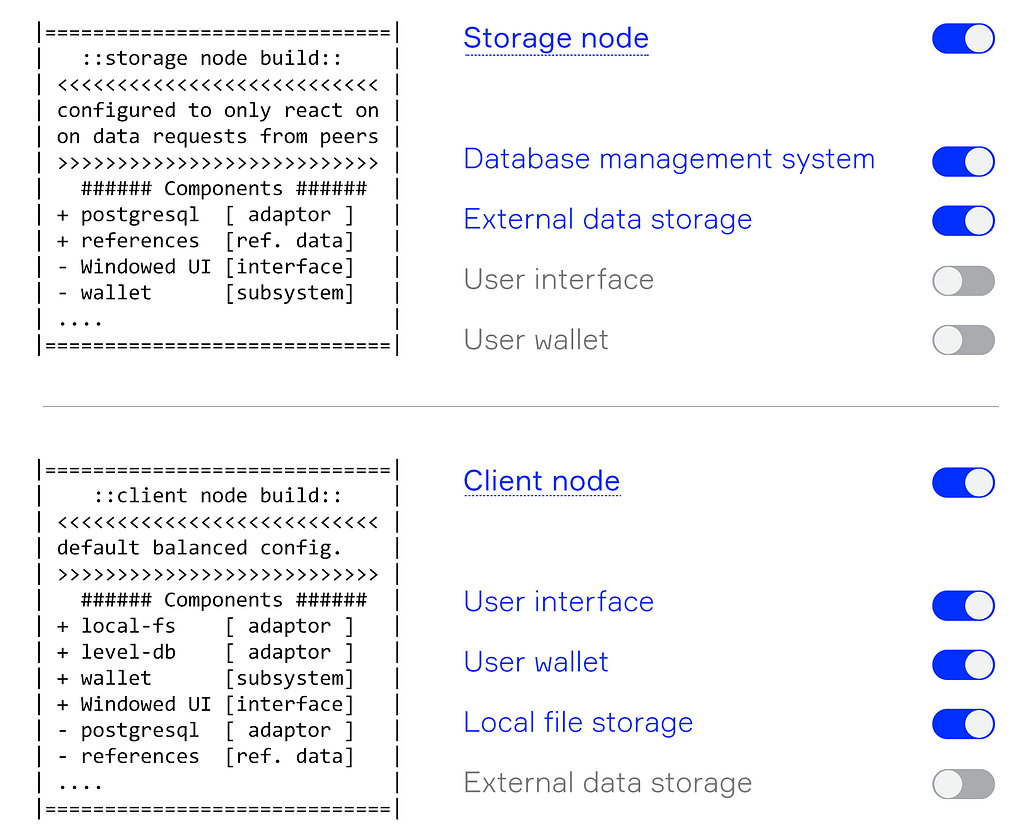
At the bottom of Figure 1 is another possible configuration for a node build. The adaptor for PostgreSQL is missing, but there is a module to expose local files to the network and the level-DB adaptor. This configuration is better suited for an ordinary user of the platform.
II. Peer-to-peer Connection. Transport level.
There are plenty of existing protocols that may easily connect nodes between each other, like TCP and WebSocket, to name a couple. The truth is, we do not care how two peers are connected. We care only about the data they send to each other.
It’s called transport-agnostic overlay network.
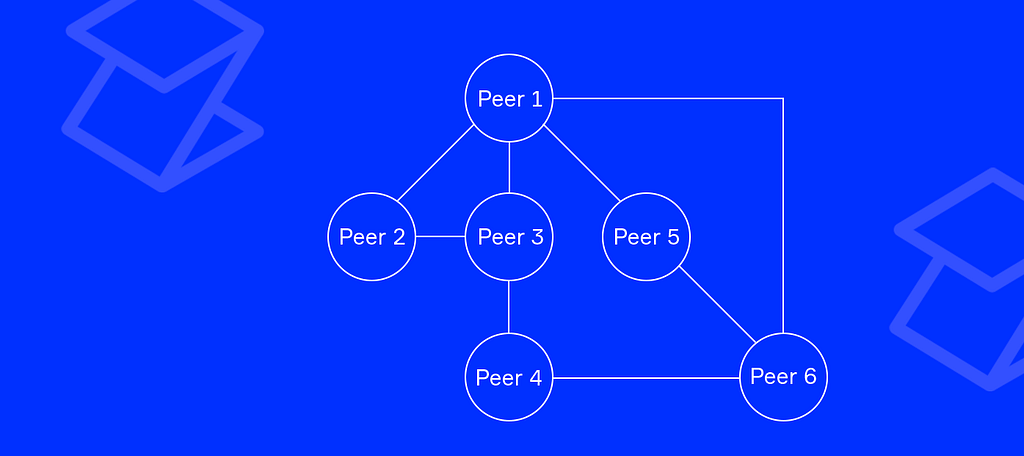
The architecture of the first network layer allows to support multiple pluggable transports and to abstract from differences between different transports. From this point, we cannot use the IP: PORT pair to reference a node — we need another means to distinguish between peers.
III. Peer Identification
This is the first time in our story where we need cryptography. Let’s create a secret and public key pair for each node in the network.

From now on, nodes in the network have their own IDs [ ID = hash {PublicKey} ]. Thanks to crypto signatures, each node can verify whether it connected to a correct node or an imposter.
We should note, that these NodeIDs are completely unrelated to user accounts.
IV. Peer Discovering and Handshaking
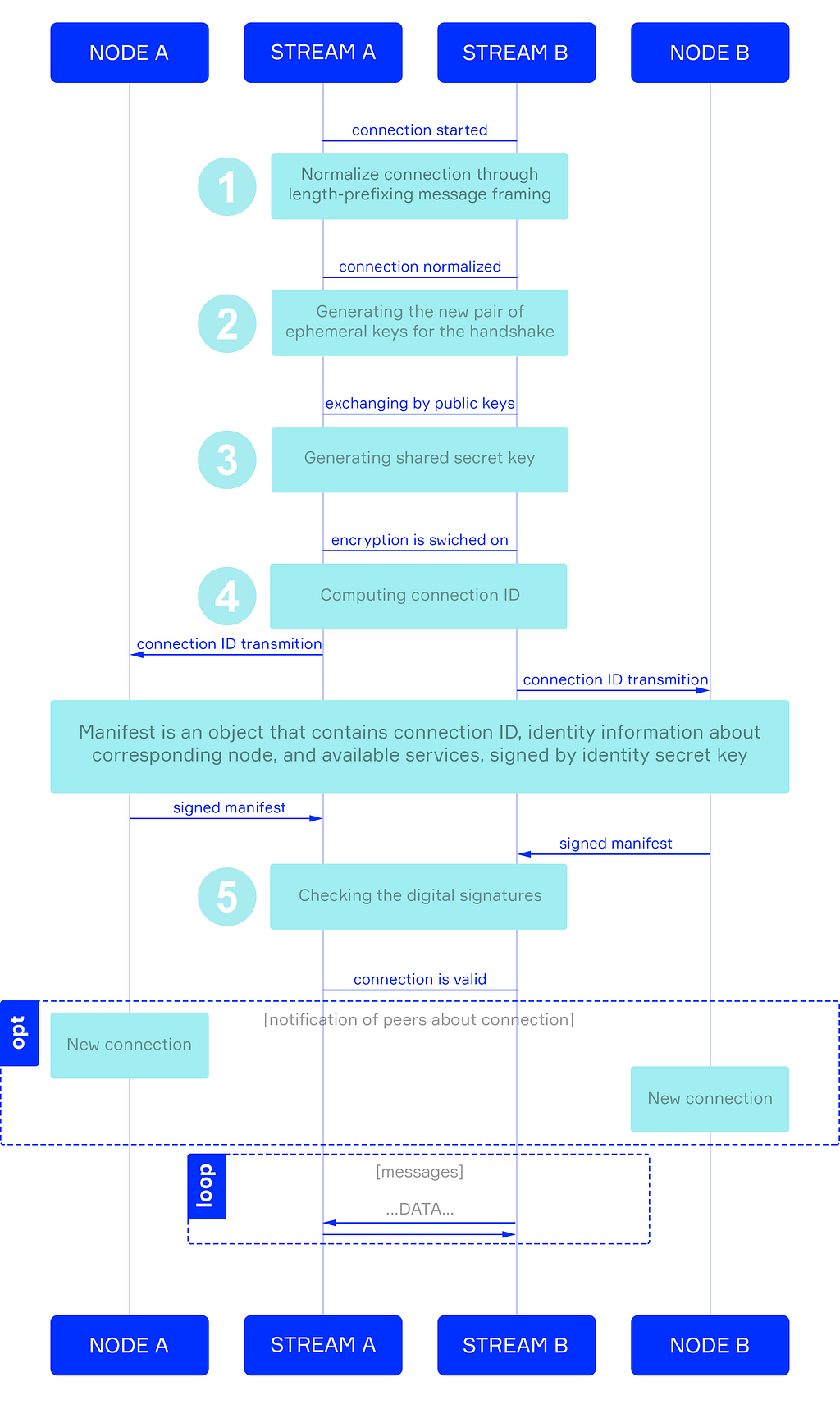
In the Zenome network, nodes prove their identity when connecting to other nodes by using a Handshaking Protocol, adapted for a distributed environment.
Figure 3 shows a diagram of the nodes handshaking, including the following stages: exchange of messages; creation, and exchange of public keys; exchange of private keys, and verification of digital signatures:
V. Peer Messaging and Events.
An event is a single piece of data that may exist on the network. When talking about an event propagating through a network, we may also call them messages.
There are many different types of messages. The simplest one is a broadcast message. Let’s study the following example.
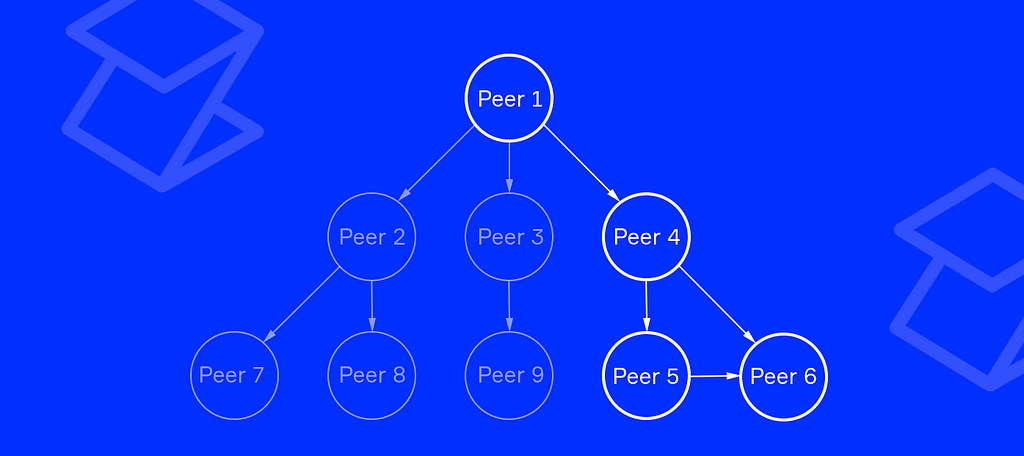
In Figure 4, we see some ordinary nodes and one huge node that belongs to a scientific organization. This organization is conducting research and would like to provide information on the network for other peers.
To do so, Peer-1 emits an event with necessary data, signs it, and broadcasts to its connecting nodes. Peer-4 processes the event when received (stores it or shows it to the user) and forwards it to the next nodes.
The process of data transfer is realized by messaging between peers. Each message contains a signature, header, and payload.
The data transfer is multiplexed, the data transfer rate corresponds to the internet stream and is primarily determined by the speed of the Internet provider.
Message structure
Figure 5 shows the structure of the message. Each message includes a header, the payload, and signatures. Compared to a conventional letter, the header is the address and name of the recipient, the payload is the content of the letter (a file), and the signatures are the sender’s name and identity confirmation.

In the JavaScript programming language, the structure of the message is as follows:

VI. Routing and Switching / Events Handling
Concerning the last example, many questions arise. Will messages travel forever on the path (Peer-4 → Peer-5 → Peer-6 → … ?) Should a node always broadcast an event? Should a node broadcast an event that has already been broadcast by this node? Is there another type of event that should be handled differently?
The architecture of the node software should naturally answer these questions. Here is how.
The software node’s architecture (the general scheme is shown in Figure 6) is implemented in such a way that the node performs the functions of the router and network card in centralized networks. Below is a diagram of the node structure and a description of its components.
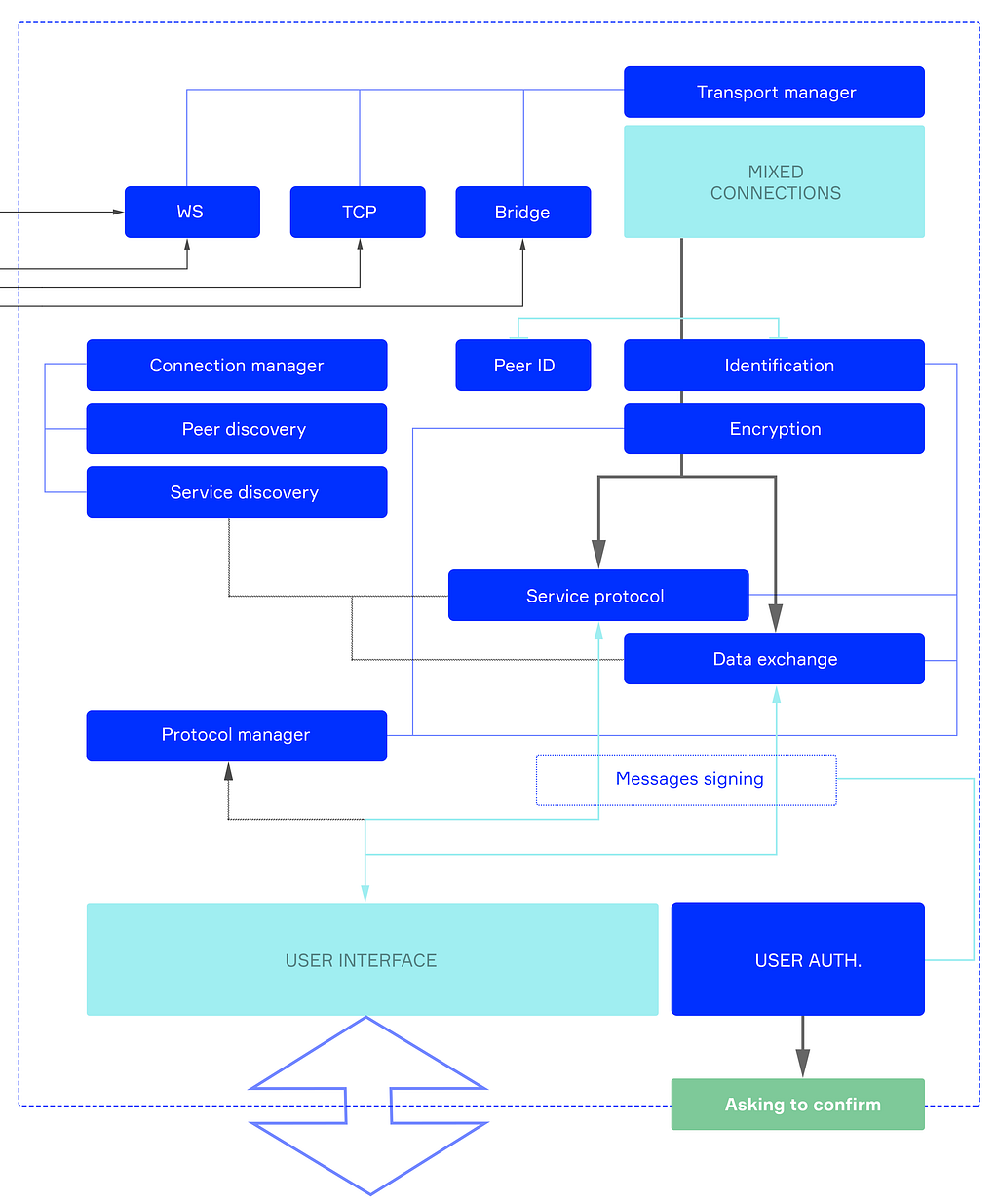
Each node at the core has two components, connected by a bus (data channel between components). These components are called South and North Bridges.
The North Bridge is responsible for actual data processing and provides all sorts of end-user interfaces through corresponding modules. When data is required, or data should be sent to the network, the North Bridge requires the South Bridge to do this.
The South Bridge is different. First, it handles network connections through a transport manager. Events from all transports will eventually come to the South Bridge input-pipeline.
Input Pipeline
Below is the operation scheme of the South Bridge, which, as a type of dispatcher or router, creates an orderly exchange of data on the network, which is critical in a decentralized environment.
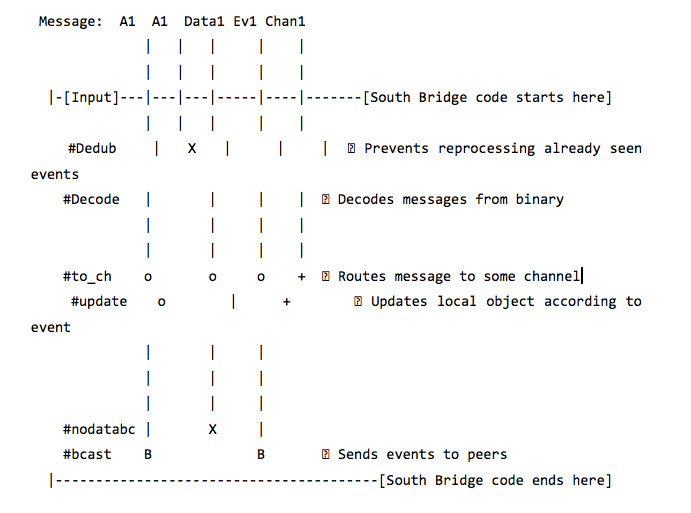
VII. Application level
Zenome Network is a general-purpose network in which the following services are possible:
● Instant Messaging Service
This service allows for the sending of text requests between peers, for example, a request to fill out a questionnaire of taste preferences or individual drug sensitivity/response. It is also possible to send mass notifications to all carriers of an “interesting genotype” to the buyer.
● File Storage Service
The file storage service allows for the loading and long-term safe storage for both genomic data and corresponding meta-information. For example, you can store raw genomic data on the Zenome Network as a “backup” with the ability to access it from any device. At the same time, data storage is organized with full privacy protection and a high-security level.
● Genomic Data Manager Service
The service allows performing various operations on genomic data in different formats: mapping of the reads, SNP calling, statistical operations (GWAS), and interpretation of data with the possibility of providing a personal genomic report.
VIII. Distributed Ledger Technology. Consensus.
Currently, the term “blockchain” has become synonymous with “distributed register technology.” The blockchain is just one of the possible topologies for recording transactions — in the form of a linear sequence (chain) of blocks. This topology has several deficiencies, including a low transaction confirmation rate. This topology creates a fundamental problem for scaling the network, which we can see in an example of Bitcoin or Ethereum.
We use a directed acyclic graph (DAG) as a distributed ledger. DAG technology implies a non-linear method of recording transactions. For a detailed review, see DAG technology advantages [8].
To achieve consensus in the Zenome network, we use the FBA (Federated Byzantine Agreement) protocol; for more detail see the Stellar project documentation [9].
The combination of such a transaction record topology and consensus achievement protocol allows us to have an almost unlimited transaction speed during testing. In a real network, we assume the speed of confirmation of one transaction will be 1–5 seconds.
The high transaction confirmation rate and, consequently, the network’s scalability are essential features for working with genomic data, medical records, and corresponding meta-information.
References
[1] https://github.com/redecentralize/alternative-internet
[3] https://icotimeline.com/wp-content/uploads/2017/07/Gene-Chain-Whitepaper.pdf
[4] https://shivom.io/files/Whitepaper.pdf
[5] https://www.genomes.io/wp-content/uploads/2018/07/The-genomes.io-Whitepaper-V-1.1.5.pdf
[6]https://www.nebulagenomics.io/assets/documents/NEBULA_whitepaper_v4.52.pdf
[7] https://zenome.io/download/whitepaper.pdf
[8] https://arxiv.org/pdf/1804.10013.pdf
[9] https://www.stellar.org/blog/stellar-consensus-protocol-proof-code/
…
Zenome Network: Distributed Genomic Internet was originally published in Zenome on Medium, where people are continuing the conversation by highlighting and responding to this story.